Arabidopsis thaliana Root Segmentation for Plant Phenotyping
Challenge Organizers: Nicolás Gaggión, Enzo Ferrante, Thomas Blein, Federico Ariel, Diego Milone, Florencia Legascue, Florencia Mammarella, Alejandra Camoirano.
Understanding plant root plastic growth is crucial to assess how different populations may respond to the same soil properties or environmental conditions and to link this developmental adaptation to their genetic background. Root system architecture (RSA) is characterized by parametrization of a grown plant, which relies on the combination of a subset of variables like main root (MR) length or density and length of the lateral roots (LRs). Root segmentation is the first step to obtain such features.
Automatic segmentation of large datasets of plant images allows the analysis of growth dynamics as well as the identification of novel time-related parameters. We have recently released an open-hardware system to capture video sequences of plant roots growing under controlled conditions. In this challenge, we call the computer vision and plant phenotyping communities to develop and benchmark new image segmentation approaches for the challenging task of plant root segmentation.
Dataset Description
For the challenge, we will present two datasets:
- Training Dataset (download, Google Drive)
- Testing Dataset: to be released on the 28th June 2021
On each dataset, we provide video sequences of arabidopsis plants growing on controlled conditions, on Petri’s boxes for 2-4 weeks. Each video sequence has one frame every 12hrs, with weakly annotations, leading to approx. 5 annotated images per video.
The video sequences are split in two different folders, according to the type of the annotation:
- Binary labels: ground truth was manually generated for 207 frames (5-7 frames per video, for a total of 34 videos). The binary labels correspond to root vs background segmentations as shown in Figure 1.
- Multi-class labels: an expert biologist segmented 129 frames (approximately 5 images per video, for a total of 26 videos). The multi-class labels correspond to main root, lateral root and background as shown in Figure 2.
Every image has a fixed resolution of 3280 x 2464 and was captured with an infrared camera, meaning the 3 color channels can be reduced to 1 without loss of information. Note that every image has a QR code associated with it. This code is used to automatically compute the pixel resolution, since its size is 1cm x 1cm.
The ground truth annotations are provided in .nii.gz format, which is a standard format for medical images that can be easily opened in python using nibabel, and visualized together with the PNG images using ITK-SNAP. Note that PNG images in ITK-SNAP can be opened as “Generic ITK Image” as file format, while the .nii.gz files can be loaded as segmentation masks and overlapped for visual inspection.
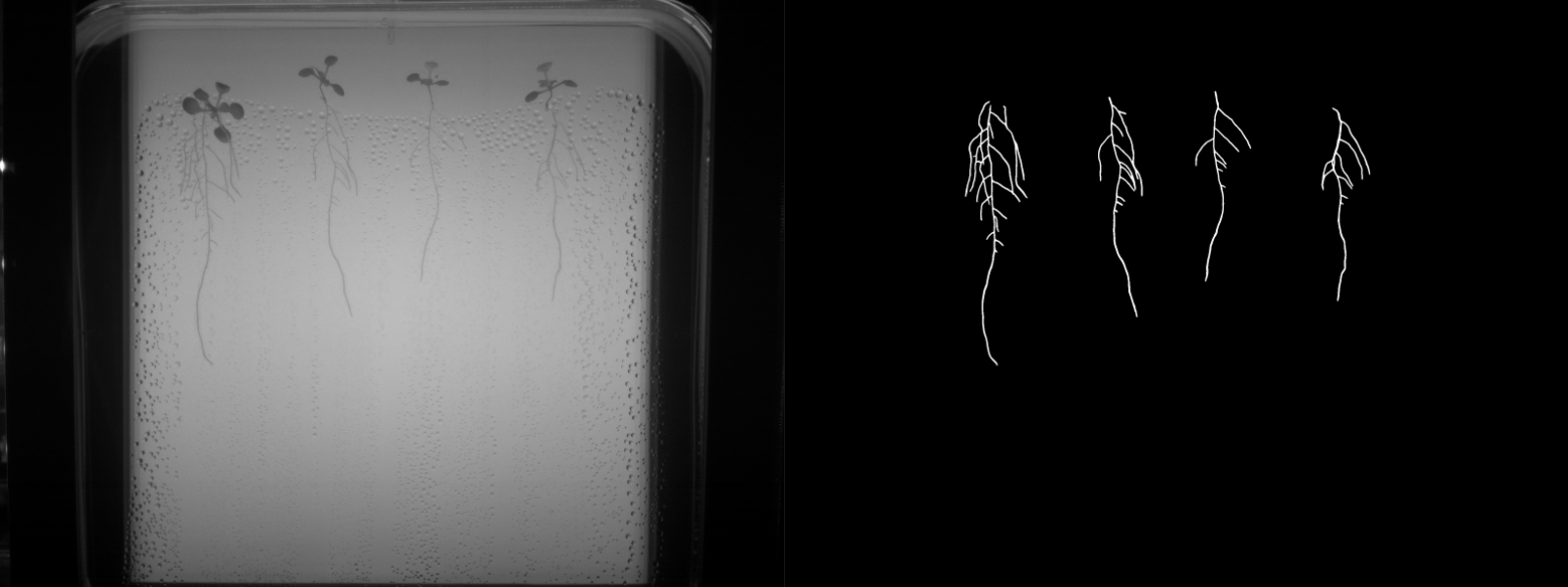
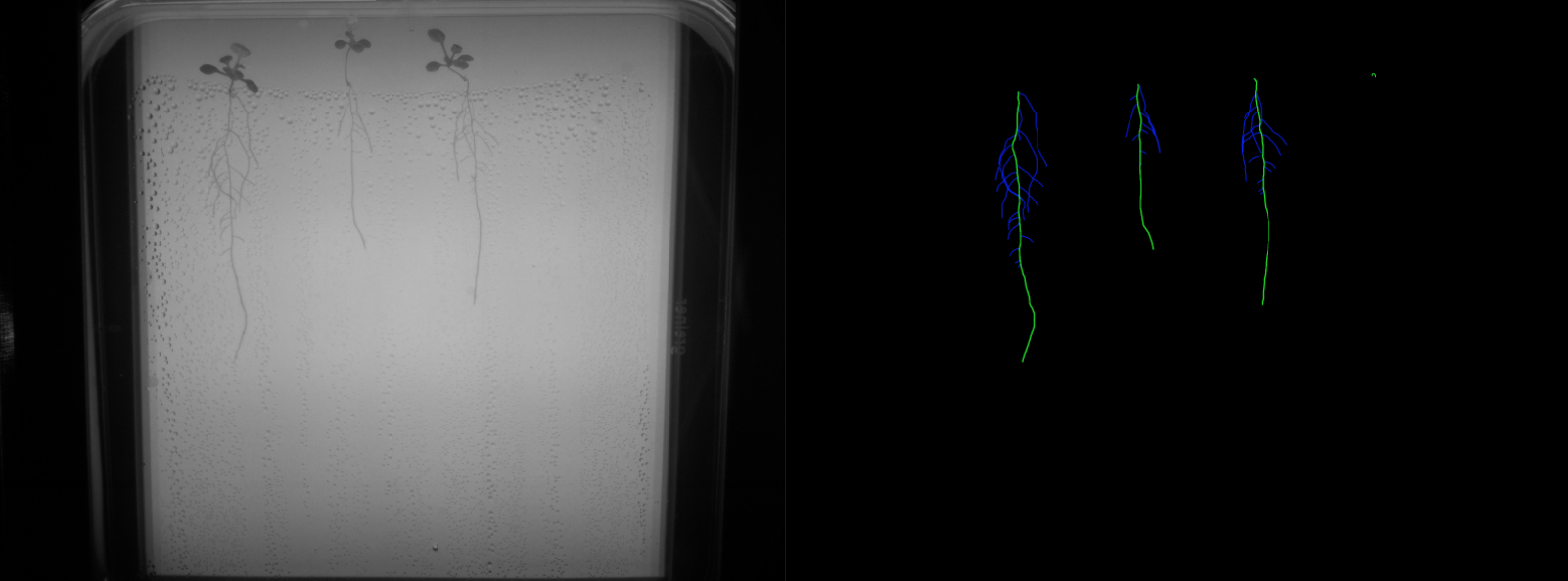
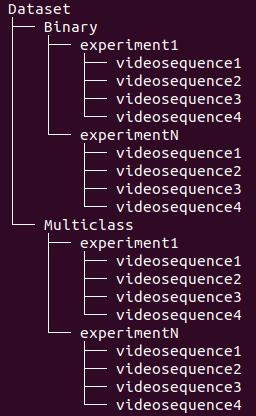
The folders are organized in the structure presented on Figure 3, where every experiment consists of a setup of our open-hardware system with upto 4 video sequences generated by its cameras.
You can use these images for research purposes only. If used in a publication, please cite the pre-print manuscript associated to ChronoRoot, the open-hardware system used to capture these images:
Gaggion, N., Ariel, F., Daric, V., Lambert, É., Legendre, S., Roulé, T., Camoirano, A., Milone, D., Crespi, M., Blein, T. and Ferrante, E., 2020. ChronoRoot: High-throughput phenotyping by deep segmentation networks reveals novel temporal parameters of plant root system architecture. Link: BiorXiv
Task Submission and Evaluation
We will benchmark the methods considering both binary and multi-class segmentation, using the ground-truth annotations produced by expert plant biologists on the dataset provided in the evaluation phase. Participants will have to submit only the multi-class segmentations, in standard .nii.gz format, for every frame of the test dataset keeping the same file name and only changing the extension. We will then derive the corresponding binary segmentations for evaluation. Results will only be evaluated in the frames where our biologists provided annotations.
As roots are thin structures, classic segmentation metrics like Dice Score may not fully reflect the quality of the results. Thus, we will evaluate participants using two types of metrics: segmentation and skeleton based metrics. The segmentation metrics will be computed based on the segmentation masks provided by the participants. These segmentations will be skeletonized using Python’s scikit-image skeletonize and then skeleton metrics will be computed.
As for the segmentation metrics we will consider: Dice Coefficient, Hausdorff distance, and number of connected components.
For the skeleton metrics we will consider skeleton completeness and correctness as discussed by Youssef et al. (2015), and main root, lateral root and total root length, counted as number of pixels from the skeleton.
We will release the testing dataset on the 28th June 2021. You can submit your data to be evaluated to rootsegchallenge@gmail.com until the 9th July 2021.
We will evaluate the metrics on the ground truth annotations of the test data, and release the final leaderboard by the end of July.
Additional Information
We believe our weakly annotated dataset with high spatial and temporal resolution may serve to evaluate fully or semi supervised approaches for plant root segmentation. Participants are encouraged to submit a full or abstract paper to the CVPPA workshop discussing the method used to segment the video sequences.
If the number and the quality of the submissions allows, we will consider to write a journal challenge paper benchmarking the different methods, where participants will be invited to contribute.
References
Youssef, R., Ricordeau, A., Sevestre-Ghalila, S., & Benazza-Benyahya, A. (2015). Evaluation protocol of skeletonization applied to grayscale curvilinear structures. In 2015 International Conference on Digital Image Computing: Techniques and Applications (DICTA) (pp. 1-6). IEEE. Link: IEEE.


